APPLICATION NOTE
Dr. Emily J Guggenheim1 and Dr. Joshua Z Rappoport2
- School of Geography Earth and Environmental Sciences, University of Birmingham, Edgbaston, Birmingham, B15 2TT, UK 2. Center for Advanced Microscopy and Nikon Imaging Centre, Feinberg School of Medicine, Northwestern University, Chicago IL, 60611, USA
Reflectance imaging for visualization of unlabelled structures using Nikon A1 and N-SIM
Reflectance imaging describes a subset of light microscopy methods that permit the formation of an intensity image from light that is back-scattered from a sample. This allows reflectance imaging to be performed alongside techniques such as fluorescence imaging, or as a standalone methodology for the investigation of a variety of labeled and unlabeled samples.
Interference Reflectance Microscopy (IRM) was the initial application of reflected light imaging1. IRM involves the formation of an image from signal that is generated by light interacting at the boundary of substrates with differing refractive indices (RI). Since this original application, a variety of different configurations have emerged leading to the generation of images based on RI mismatches that are naturally occurring throughout cell and tissues samples. However, contrast can also be artificially induced by the introduction of materials with high RI and optical density, analogous to fluorescent labeling.
The Nikon A1 point-scanning confocal and the Nikon N-SIM super-resolution structured illumination system are both capable of imaging in reflectance mode; each technique offering different advantages and disadvantages for critical investigations into reflectant substrates, such as nanoparticles (NPs), which have previously been discussed2,3. Other modalities can also benefit from the use of reflectance configuration to provide structural information, including multiphoton in vivo investigations4.
Reflectance imaging: introduction
Contrast is generated in microscopy images through the interaction of light with different specimen components. In traditional fluorescence microscopy this signal is generated through the emission of light from excited fluorophores, conjugated to molecules of interest. However fluorophores can be difficult to attach, may alter physicochemical properties and cellular processing of labeled substrate, can detach during experiments, and are subject to photobleaching5,6.
In transmission and reflectance imaging contrast is generated by the interaction of light at boundaries present within the sample.
These boundaries are present at the regional interfaces where RI differs; in transmitted light microscopy it is the light that is transmitted through that is collected, whereas in reflectance microscopy it is the back-scattered light that forms the image. Reflectance offers several advantages in terms of sample interrogation:
- No labels are required to visualize various components within the sample as with fluorescence imaging
- Photobleaching does not occur as in fluorescence imaging
- The Signal-to-Noise Ratio (SNR) of objects of interest is greatly enhanced compared to transmission microscopy
Therefore reflectance imaging becomes the obvious choice for the investigation of samples that contain objects of interest with substantially different RI’s or optical densities to their surroundings – such as the imaging of intracellular metallic NPs2,3,7,8.
Reflectance imaging using the Nikon A1 confocal system
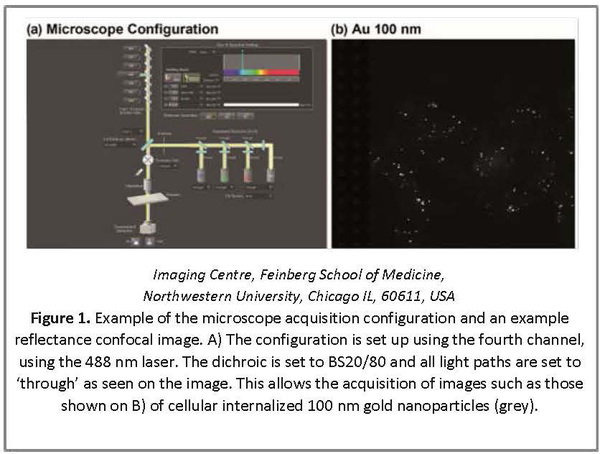
One of the initial limitations of reflectance imaging is the high level of background present in highly scattering environments, such as cells. Confocal microscopy provides a means to increase both SNR and resolution compared to epi-illumination in reflectance mode by filtering out-of-focus light through the conjugate pinhole system. This also permits optical sectioning. Confocal reflectance images can be acquired using a confocal microscope such as the Nikon A1 by simply changing the dichroic to the transmitted / reflectance mirror (BS20/80) and set up of the 4th channel with the laser wavelength of choice (e.g. 488 nm) (Figure 1) (Guggenheim et al., 2016).
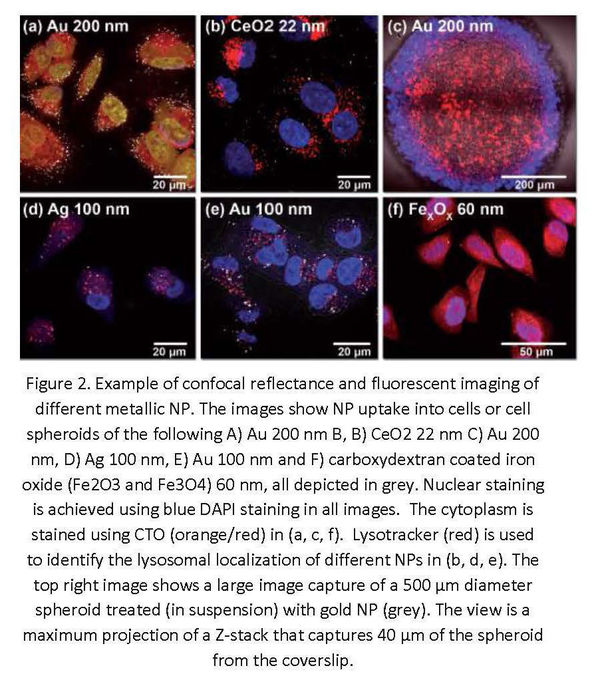
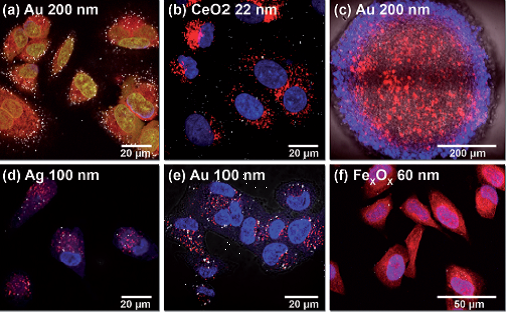
RCM can provide semi-quantitative results, particularly beneficial within the field of nanoresearch when combined with automated processing and analysis to increase the throughput of studies2,3. However, in order to get absolute quantitative results, efforts have been made to perform correlative experiments by the incorporation of modalities such as Transmission Electron Microscopy (TEM) and Mass Spectrometry (MS) into workflows2,11.
Reflectance imaging using the Nikon N-SIM super-resolution system
Despite the advantages that confocal reflectance offers, the fundamental restrictions on resolution that govern conventional light imaging modalities still apply. Therefore increasing effort is being placed on the ability to acquire images with resolutions that surpass the diffraction limit of light, collectively termed super-resolution techniques. Structured Illumination Microscopy (SIM) can increase the spatial resolution two-fold with relatively small loss in temporal resolution, rendering it advantageous for live cell imaging12. Despite the widespread use of SIM in fluorescence imaging, the use of SIM in reflectance mode is not often documented.
In a traditional Nikon N-SIM acquisition, a filter cube is present within the light path consisting of an excitation filter, emission (or barrier) filter and a dichroic mirror. This allows light emitted from the specimen to pass through and reach the detector, while also blocking the detection of the excitation light. When performing Reflectance SIM (R-SIM), the filter cube is replaced with a half mirror that permits the transmission of the light reflected from a sample at the same wavelength as the incident light. In R-SIM the sample is illuminated in the same manner as for fluorescent SIM, whereby a grating is used to structure the incident light allowing acquisition of several images that can then be reconstructed in the Fourier domain to yield the super-resolution image12. This method has been applied to the investigation of intracellular NPs using both custom and commercial microscopes2,7.
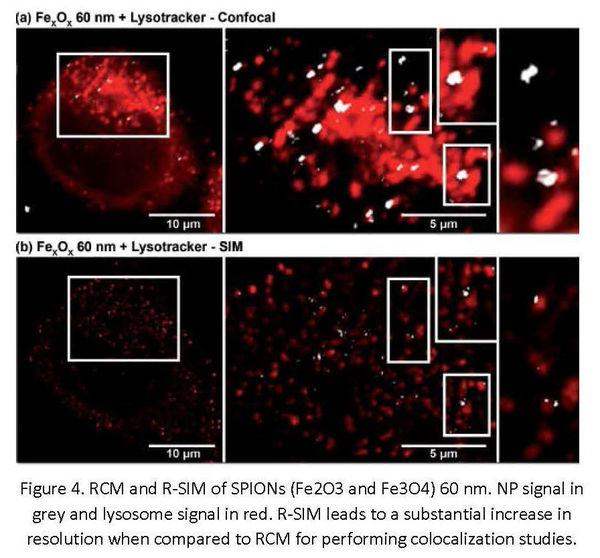
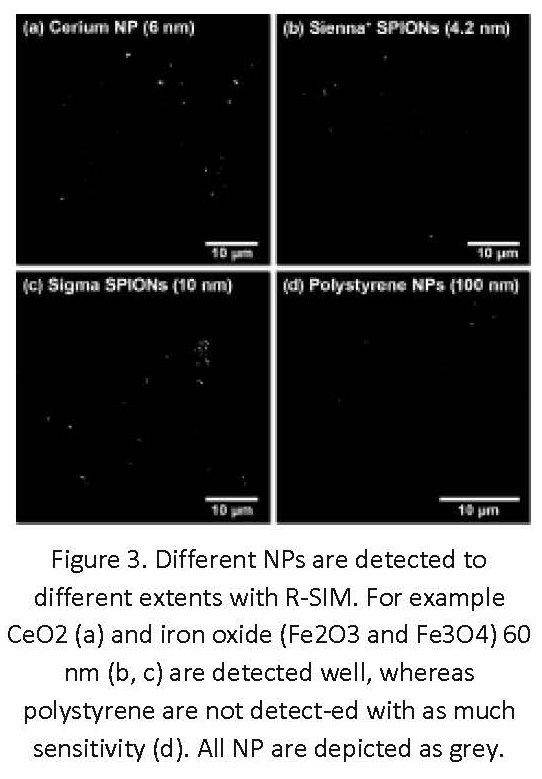
The ability to resolve structures separated by less than the diffraction limit by label-free light microscopy offers significant advantages for localizing the NP clusters with increased precision and accuracy. NPs with varied compositions appear to be detected, albeit with different efficiency using R-SIM (Figure 3). For example, superparamagnetic iron oxide nanoparticles (SPIONs) appear to be better detected by RCM than R-SIM (Figure 4)2. Lower detection with R-SIM is a result of the wide-field illumination method used, and therefore high levels of background scattering. R-SIM can therefore be a particularly useful source of additional resolution and information within biological investigations, due to the minimal sample preparation necessary and, like with confocal, ease of combination with fluorescent labeling. Therefore R-SIM with the Nikon N-SIM instrument can offer more certainty in studies such as colocalization assessment of NPs and labeled structures (Figure 4). The sensitivity and specificity of reflectance methods can be confirmed by using correlative workflows that include TEM, as done in previous studies2.
Conclusions
Different illumination and scatter collection configurations can be employed to perform reflectance imaging; these include interferometry, confocal, evanescent wave, structured and oblique angle illumination. This facilitates a wide range of possible studies, particularly within the field of nanoresearch, where reflectance investigations will be instrumental in determining the safety and efficacy profiles of NPs for various applications. As different components of cells and tissue have different RIs, reflectance imaging can be advantageous for probing the structure and organization of biological samples. The power of reflectance for providing additional and complimentary structural information is particularly pronounced in the field of in vivo imaging, contextualizing existing diagnostic and monitoring strategies.
References
- Curtis, A. S. (1964) ‘The Mechanism of Adhesion of Cells to Glass. a Study by Interference Reflection Microscopy’, The Journal of cell biology. UNITED STATES, 20, pp. 199–215.
- Guggenheim, E. J. et al. (2016) ‘Comparison of Confocal and Super-Resolution Reflectance Imaging of Metal Oxide Nanoparticles’, PLOS ONE. Edited by V. E. Degtyar. Public Library of Science, 11(10), p. e0159980. doi: 10.1371/ journal.pone.0159980.
- Guggenheim, E. J., Lynch, I. and Rappoport, J. Z. (2017) ‘Imaging In focus: Reflected light imaging: Techniques and applications’, The International Journal of Biochemistry & Cell Biology, 83, pp. 65–70. doi: 10.1016/j.biocel.2016.12.008.
- Costantini, I. et al. (2016) ‘Combination of two-photon fluorescence microscopy and label-free near-infrared reflectance: a new complementary approach for brain imaging’, in Biomedical Optics 2016. Washington, D.C.: OSA, p. JW3A.23. doi: 10.1364/CANCER.2016.JW3A.23.
- Sugden, J. K. (2004) ‘Photochemistry of dyes and fluorochromes used in biology and medicine: some physicochemical background and current applications.’, Biotechnic & histochemistry : official publication of the Biological Stain Commission, 79(2), pp. 71–90. doi: 10.1080/10520290412331292400.
- Quinn, M. K. et al. (2015) ‘How fluorescent labelling alters the solution behaviour of proteins.’, Physical chemistry chemical physics : PCCP. The Royal Society of Chemistry, 17(46), pp. 31177–87. doi: 10.1039/c5cp04463d.
- Chang, B.-J. et al. (2011) ‘Subdiffraction scattered light imaging of gold nanoparticles using structured illumination’, Optics Letters. Optical Society of America, 36(24), pp. 4773–4775.
- Mazzolini, J. et al. (2016) ‘Protein corona modulates uptake and toxicity of nanoceria via clathrin-mediated endocytosis e’, The Biological Bulletin.
- Gáspár, I. and Szabad, J. (2009) ‘In vivo analysis of MT-based vesicle transport by confocal reflection microscopy.’, Cell motility and the cytoskeleton, 66(2), pp. 68–7 doi: 10.1002/cm.20334.
- Stensberg, M. C. et al. (2014) ‘Silver nanoparticle-specific mitotoxicity in Daphnia magna.’, Nanotoxicology, 8(8), pp. 833–42. doi: 3109/ 17435390.2013.832430.
- Kim, C. S. et al. (2015) ‘Cellular imaging of endosome entrapped small gold nanoparticles’, MethodsX, 2, pp. 306–315. doi: 10.1016/j.mex.2015.06.001.
- Gustafsson, M. G. (2000) ‘Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy.’, Journal of microscopy, 198(Pt 2), pp. 82–7. Available at: http://www.ncbi.nlm.nih.gov/ pubmed/10810003 (Accessed: 3 May 2016).